AI makes key protein research a million times faster
Computers can be trained to figure out how these key building blocks of life work, a Harvard researcher finds.
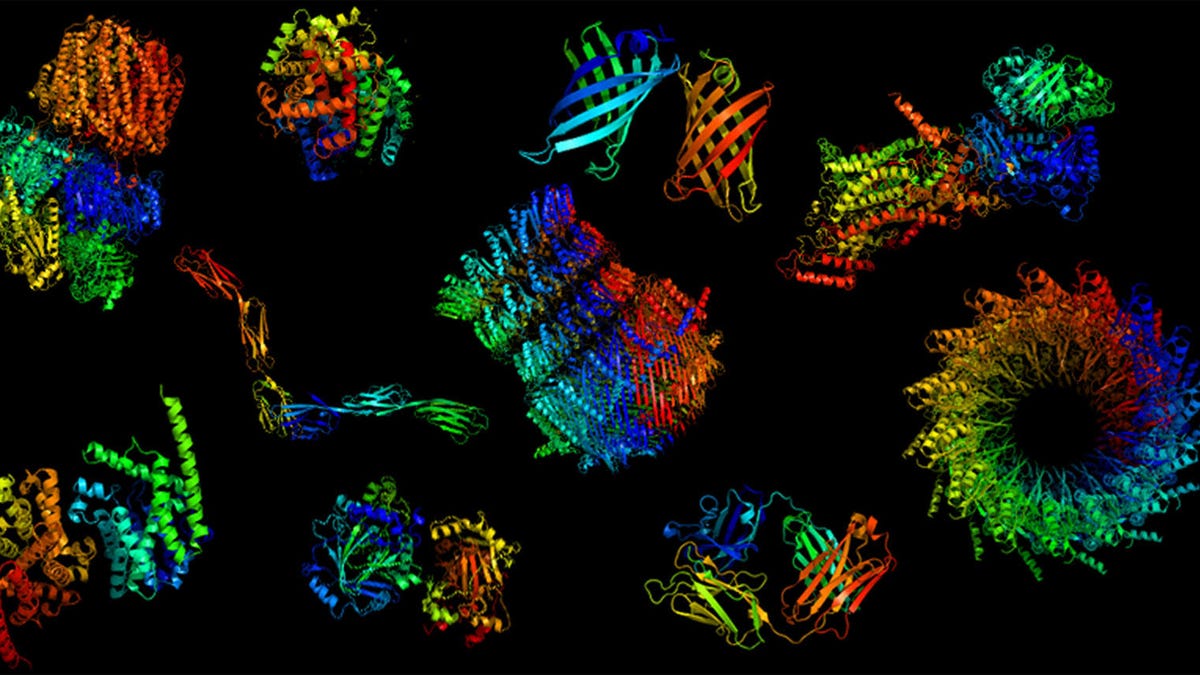
Proteins, like these shown in 3D computer models, are highly complex biological molecules.
Protein folding has been a famously difficult computing problem for decades: How do you figure out the exact structure of these massive molecules that our DNA defines? Now artificial intelligence is getting us to the answer a lot faster.
Harvard Medical School biologist Mohammed AlQuraishi has used the latest machine learning technology to detect structural patterns in well-understood proteins, then apply that to other proteins.
The results, while not precise enough for protein folding applications like discovering new drugs, is at least a million times faster than conventional computing techniques. And this is just a first crack at a technology that can be improved and combined with other modeling techniques.
It's an illustration that AI, though fraught with fears about effects like abetting police states or wiping out human jobs, has the potential to improve medicine, among other things.
Mohammed AlQuraishi from Harvard Medical School developed an AI technique to predict how crucial biological molecules called proteins form. As his model improves, the colorful prediction gradually comes closer to the actual protein structure shown in gray.
"We now have a whole new vista from which to explore protein folding," AlQuraishi said in a statement Wednesday. "We've just begun to scratch the surface."
AI today most often refers to neural network technology loosely based on human brains, and it's revolutionized everything from voice commands and facial recognition to debugging software and turning on windshield wipers. AI models learn patterns from real-world training data, an approach that means humans don't have to make specific instructions like trying to define what it sounds like when somebody says "Alexa, what's the weather today?"
In humans and any other form of life on Earth, DNA strands contain instructions on how to assemble amino acids into the long strings that become proteins. The laws of physics determine exactly how those strings collapse into tight bundles, with the resulting surface structures critical to protein interactions inside cells.
But modeling exactly how that will happen inside a computer quickly gets difficult for bigger proteins. That means it's hard to understand what's going on with proteins. AlQuraishi, though, believes the AI technique could not only help this understanding but also potentially be used to engineer new proteins that perform a specific job.
AlQuraishi's results were published Wednesday in the journal Cell Systems.

