How the antibiotic chloramphenicol causes damage to eukaryotes
A group of scientists from Japan led by Professor Takashi Kamakura of Tokyo University of Science has demonstrated the molecular and cellular basis of the toxic effects of the antibiotic chloramphenicol on eukaryotic cells. Concluding their study published in Scientific Reports, they write, "Identification of the molecular target of chloramphenicol may lead to better elucidation and resolution of its side effects in humans."
Antibiotics are often the treatment of choice for bacterial or fungal infections. Chloramphenicol is an example of a broad-spectrum antibiotic that's active against most bacteria and is widely used in human and veterinary medicine. However, it has varied side effects, including human aplastic anemia, bone marrow suppression, and gray baby syndrome, which limit its prescription. A drug's side effects are usually thought to be due to its interaction with entities other than the target molecules. The causes of the side effects are not fully understood, but the Japanese researchers have now demonstrated one of the possible mechanisms of the effect of chloramphenicol on eukaryotic cells. They write, "High doses of chloramphenicol have been known to cause mitochondrial damage in eukaryotes ... our study shows that drugs have novel secondary targets in eukaryotes."
As the model eukaryotic organism for the study, they used the rice blast fungus Pyricularia (Magnaporthe) oryzae. Regarding the significance of this model organism, Professor Kamakura says, "The rice blast fungus is an economically important pathogen that causes destructive disease in rice, which is the staple food of Japan. Thus, from the perspective of food security, it is important to understand the infection strategy of this pathogen and implement ways to control the damage to crops. The genome of this fungus has considerably less 'junk' DNA—the 'noncoding' part of the genome that does not code for any functional proteins. This, along with the low number of targetable functional proteins, makes the rice blast fungus an ideal model for the screening of novel molecular targets of drugs."
The rice blast fungus uses a peculiar structure called the appressorium, which functions in surface recognition, binding and subsequent infection. The fungus infects the plant using a three-cell structure called the conidium, which has a germ tube with the appressorium at the end. The appressorium initiates contact with the host (plant) cell and is crucial in the infection cycle. This entire process is known to be dependent on the mitotic division of the single-cell germ tube and cell differentiation. Given the fact that cell differentiation depends on several molecular components, these molecules can also be targets of several drugs. The researchers exploited this principle.
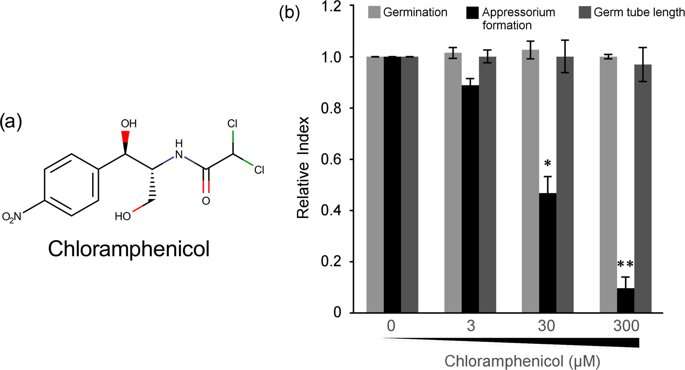
The researchers first exposed a suspension of the fungal conidia to different concentrations of chloramphenicol. After six hours, they found via microscopic observation that the germination of the conidia or the length of the germ tube remained unaffected, but the percentage of appressorium formation significantly dwindled. Professor Kamakura's team thus concluded that chloramphenicol specifically targets appressorium formation in the rice blast fungus, which is a novel finding.
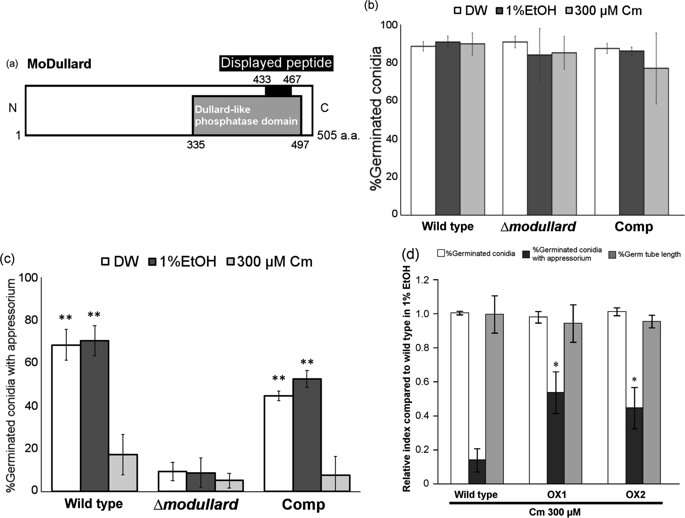
Next, they screened for chloramphenicol targets in the entire genome of the rice blast fungus. Of the 86 peptides that were targeted, 14 were known rice blast fungus peptides, and only two were functionally active proteins (proteins from the coding region of a gene). One of these was extremely similar to a highly conserved eukaryotic protein called Dullard, and the researchers termed this MoDullard. The Dullard protein is known to play a role in cell differentiation in eukaryotes. The researchers found that MoDullard was expressed the most at the appressorium formation stage. They also performed functional analysis by knocking out the MoDullard gene, and found that mutants with no MoDullard were not able to produce the appressorium, but were also resistant to chloramphenicol. This confirmed that chloramphenicol targets this protein.
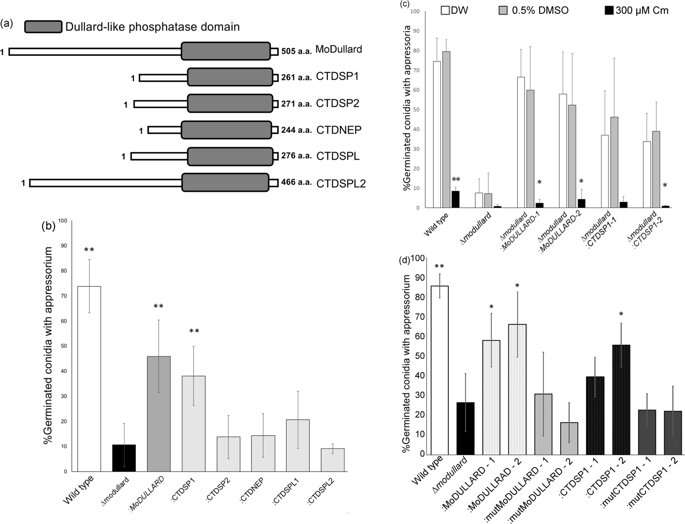
Finally, to determine the target in humans, they identified five candidate peptides from the human genome that bore resemblance to MoDullard. They isolated the gene for each of these peptides and transferred it into the MoDullard-knockout mutant. They found that one of the peptides, called CTDSP1, complemented the lost function of MoDullard in the mutant cells—meaning that the cells with CTDSP1 introduced were able to produce appressorium. Chloramphenicol was again found to block the formation of this appressorium, which led the researchers to confirm that CTDSP1 is indeed a target of chloramphenicol in humans.
To put these results into perspective, Professor Kamakura says, "A more detailed analysis of the mechanism of inhibition of appressorium formation, revealed for the first time here, may lead to the development of new control methods for the rice blast fungus. Also, drug screening using the rice blast fungus may lead to the elucidation of yet-unknown side effects or drug repositioning to discover novel effects of existing drugs whose safety profiles have been confirmed."
More information: Akihito Nozaka et al. Chloramphenicol inhibits eukaryotic Ser/Thr phosphatase and infection-specific cell differentiation in the rice blast fungus, Scientific Reports (2019). DOI: 10.1038/s41598-019-41039-x
Journal information: Scientific Reports
Provided by Tokyo University of Science