New portable DNA sequencer quickly and accurately diagnoses wheat viruses
Blasts cause significant loses in wheat crops. Recently Bangladesh was devastated by an invasion of South American races of wheat blast fungus, which occurred for the first time in the country in 2016. The disease spread to an estimated 15,000 hectares (16% of cultivated wheat area in the country) and resulted in yield losses as high as 100%.
Diagnosis of crop disease is crucial but traditional methods rely on the expertise of pathologists, who in turn rely on the physical appearance of disease symptoms, which can be similar to damage caused by other factors, such as nutrient deficiencies or environmental elements. Pathologists also experience difficulty detecting coinfections and pathogens that do not infect aerial parts of the plant.
Furthermore, there is no existing method for rapidly identifying unknown pathogens during an outbreak, as was made clear during the wheat blast fungus outbreak in Bangladesh.
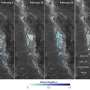
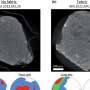
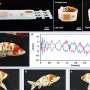
A group of scientists in Kansas have developed a new technology that makes it possible to rapidly identify viruses in wheat fields with a significantly higher accuracy. They collected four wheat samples from western Kansas and used a new harmonica-sized DNA sequencer and a computer program to quickly detect three different viruses in the samples. Furthermore, their results suggested that the samples contained a new virus strain.
These scientists, based at the USDA-ARS and Kansas State University, are now working on improving the technique so that it can be used in field applications. Their research, described in "Wheat Virus Identification Within Infected Tissue Using Nanopore Sequencing Technology" published in the September issue of Plant Disease, is the first report of using the new portable DNA sequencing technology for wheat virus identification. These results have broad application to plant and animal disease identification and field diagnostics technology.
More information: John P. Fellers et al, Wheat Virus Identification Within Infected Tissue Using Nanopore Sequencing Technology, Plant Disease (2019). DOI: 10.1094/PDIS-09-18-1700-RE
Provided by American Phytopathological Society