This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Scientists use serial crystallography to enhance 'molecular movies' and shed light on antibiotic resistance
Eadweard Muybridge's electrifying photos of a galloping horse set the world on fire when he created the precursor to what became motion pictures. For today's scientists, a new upgrade to one of the world's most powerful hard X-ray light sources could improve the way molecular movies are made. These could reveal hidden secrets of different chemicals, potentially paving the way for new treatments and pharmaceuticals.
Scientists typically use different forms of a technique called crystallography to reconstruct the molecular structure of proteins. Researchers at the U.S. Department of Energy's (DOE) Argonne National Laboratory have now used and expanded a new method called serial crystallography, developed previously at X-ray free-electron laser facilities. A paper based on the study appeared Nature Communications.
Combining serial crystallography with observations over short time scales (from a tenth to a hundredth of a second) allows scientists to detect real-time changes in the shape of proteins and bound molecules during chemical reactions. The technique offers a unique advantage over previous forms of crystallography, as individual crystals can be smaller and they only need to be illuminated with X-ray beams once, and for a short period of time.
The upcoming upgrade to Argonne's Advanced Photon Source (APS), a DOE Office of Science user facility at Argonne, will create X-ray beams that are up to 500 times brighter than those currently generated at the facility. This will allow for serial crystallography to be more broadly available at the APS, said Argonne distinguished fellow Andrzej Joachimiak, who is also the director of the Structural Biology Center (SBC) at the APS and a professor at the University of Chicago.
"Serial crystallography is really in its infancy and has largely been the purview of specialized facilities with free-electron lasers," said Joachimiak. "With the APS Upgrade, we will have the ability to study all sorts of reactions as they happen, especially for biological systems."
In a recent experiment in conjunction with researchers at the University of Chicago, Joachimiak used serial crystallography to examine the reaction of an antibiotic drug and an enzyme isolated from a drug-resistant pathogen. The result could help give researchers a better idea of the molecular mechanisms that enable certain bacteria to gain antibiotic resistance.
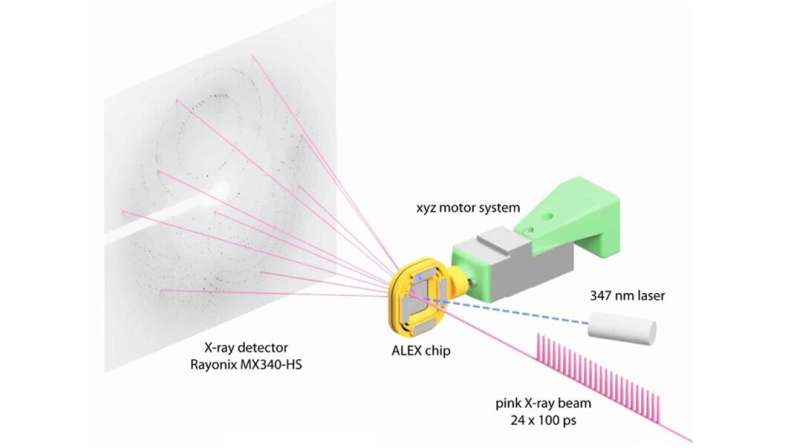
The research team made use of SBC at beamline 19-ID and the specialized capabilities of the BioCARS beamline at 14-ID, managed by the University of Chicago. According to Vukica Srajer, a BioCARS research beamline scientist and a co-author on the paper, the beamline is one of a few in the world that can perform serial crystallography on incredibly short time scales. Researchers used BioCARS to perform time-resolved serial crystallography.
By doing an X-ray scan of a plastic chip that contained a suspension of thousands of individual protein microcrystals, Joachimiak and his colleagues were able to quickly and accurately reconstruct several protein structures. The researchers used a "pump-probe" technique in which they shined ultraviolet light on the sample to initiate a reaction. They then used the X-ray beam to observe the result at different time points.
"Previous attempts at doing this kind of crystallography would destroy the crystals before we could get the complete data," Joachimiak said. "Because we're only shining the X-ray beams on each particular crystal for a very short period of time, we can get over 40,000 images from one chip. This dramatically speeds up our crystallography efforts and gives us the ability to see into protein mechanisms over several time scales we'd never before been able to resolve."
The advantage of doing serial crystallography, according to Joachimiak, is that it allows scientists to observe changes in the protein's structure as they happen. In the case of an enzyme, it also gives scientists the ability to look at how the enzyme's active site interacts with another molecule, or substrate.
In the study, Joachimiak and his colleagues looked at a protein-enzyme complex called a beta-lactamase, which gives certain pathogens antibiotic resistance. With serial crystallography, the researchers were able to notice a buildup of zinc atoms that triggered the enzyme to break through the antibiotic molecule.
"It's as if you were trying to open a jar with a stuck lid," Joachimiak said. "You keep twisting and twisting and it doesn't move initially, until ultimately it suddenly gives way."
According to Joachimiak, a water molecule becomes activated by zinc ions to break the bond of the antibiotic. "Serial crystallography shows us exactly when the zinc makes the reaction happen," he said. "You can watch it happen in real time."
Mateusz Wilamowski, a researcher at Jagellonian University in Poland and a former postdoctoral researcher at the University of Chicago, who also helped perform the research, said that the ability to resolve the dynamics of this particular class of molecules could have broad-reaching implications. "There are many other proteins like this that rely on similar mechanisms," he said. "Nobody has been able to study the intermediate transitions of the molecule that we were able to visualize."
Regular protein crystallography does not allow for the creation of these molecular movies because scientists can only collect a handful of images before destroying the crystal, Joachimiak explained. "It's truly a revolutionary technique that will have a huge impact on how we can observe and ultimately design better drugs," he said.
Wilamowski also believes that the results will help enable intelligent drug design, as future research could pair the APS Upgrade with quantum mechanical calculations to improve already existing molecules.
More information: M. Wilamowski et al, Time-resolved β-lactam cleavage by L1 metallo-β-lactamase, Nature Communications (2022). DOI: 10.1038/s41467-022-35029-3
Journal information: Nature Communications
Provided by Argonne National Laboratory





















